The Role of NGS Sequencing Database
In light of the accelerated advancements in technology, especially the meteoric evolution within the domain of bioinformatics, the significance of Next-Generation Sequencing (NGS) technologies as an essential instrument in life science research has markedly increased. Distinguished by its high-throughput, efficiency, and unerring accuracy, NGS offers unparalleled prospects in exploring the intricacies of life sciences. Given this context, the role of NGS sequence databases has increasingly assumed a pivotal role. These repositories have matured into an irreplaceable cornerstone within the field of life sciences, progressively shaping and expanding our comprehensive understanding of the complex tapestry that is biological systems.
NGS database stands as a comprehensive repository integrating a vast expanse of sequencing data. Within its confines, it amalgamates the diligent endeavors and research accomplishments of scientists worldwide, furnishing researchers with a rich and invaluable trove of experimental data resources. Facilitated by the NGS sequencing database, investigators gain facile access to sequencing data from diverse biological specimens, thereby enabling the unraveling of underlying biological principles and mysteries concealed within the data.
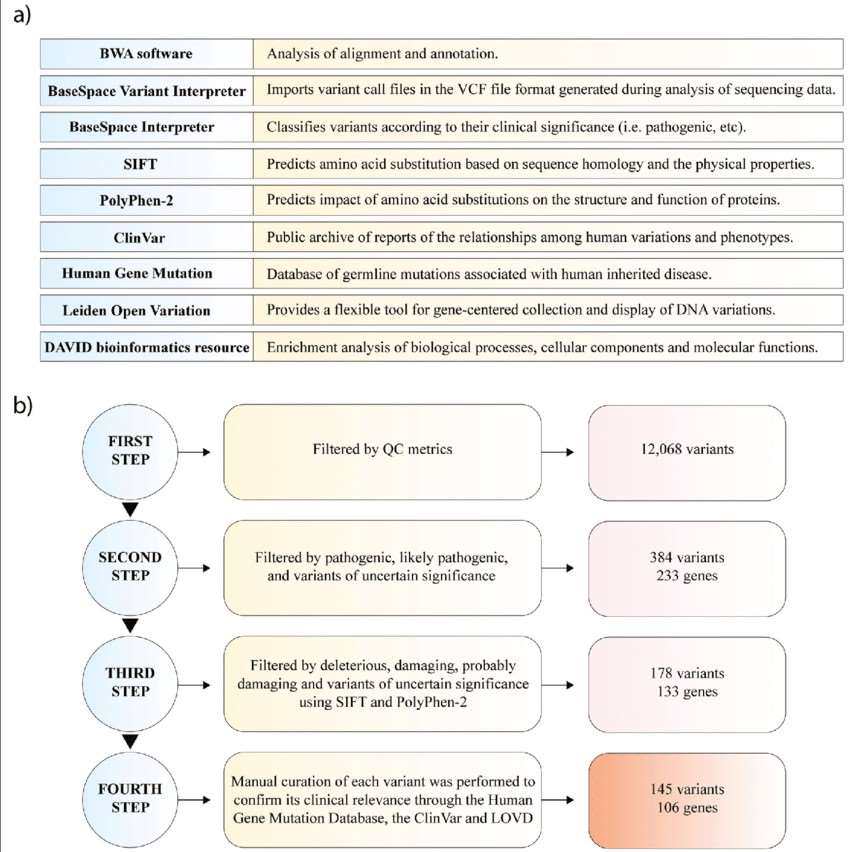
Next-generation sequencing analysis. a Functions of software and databases used for NGS analysis. b Pipeline of genomic variant analysis. (Andrés López-Cortés et al,. 2020)
The role of NGS databases manifests across several dimensions:
Data Storage and Management:
NGS sequencing databases possess robust capabilities for data storage and management. They proficiently organize vast quantities of sequencing data in an efficient and structured manner, offering a multitude of convenient querying and retrieval tools that enable researchers to swiftly locate the desired data. Furthermore, these databases feature data backup and recovery mechanisms, ensuring the security and reliability of the data.
Data Analysis and Exploration:
In addition to furnishing abundant data resources, NGS databases are equipped with potent data analysis tools. Researchers harness these tools to conduct in-depth analysis and exploration of sequencing data, unveiling biological phenomena such as gene variations and expression regulation. These insights provide scientific foundations for disease diagnosis, drug development, and personalized therapies.
Data Sharing and Exchange:
Data sharing and exchange are pivotal in the realm of NGS databases, acting as expansive platforms for collaborative endeavors among researchers. Here, scientists have the opportunity to contribute their own sequencing data, thus facilitating the dissemination of their research findings to peers within the scientific community. Concurrently, researchers gain access to data contributed by others, effectively broadening their research horizons and fostering interdisciplinary collaboration. This collaborative model of data sharing and exchange is instrumental in propelling rapid advancements in life science research, driving academic collaboration and innovation forward.
Decision Support and Assistance:
NGS databases play a crucial role in decision support and assistance. They provide policymakers, clinicians, and researchers with data support regarding disease mechanisms, drug development, and treatment efficacy, aiding them in making more scientifically informed and rational decisions.
In summary, NGS sequencing databases are indispensable in life science research. They not only offer vast sequencing data resources but also encompass various functionalities such as data storage and management, data analysis and exploration, data sharing and exchange, as well as decision support and assistance. With the continuous advancement and refinement of NGS technology, the significance of NGS sequencing databases will be further accentuated, making greater contributions to the progress and innovation of life science research. This paper aims to systematically outline and elucidate the commonly used database resources in interpreting NGS sequencing reports.
Population Databases
RefSeqGene Database
The RefSeqGene Database, accessible at http://www.ncbi.nlm.nih.gov/refseq/rsg, stands as an openly available repository housing nucleotide sequences (DNA, RNA) along with their corresponding protein products. Conceived and developed by the National Center for Biotechnology Information (NCBI) in 2000, it retains responsibility for the curation and upkeep of this invaluable resource.
Distinguished by its taxonomic diversity, non-redundancy, meticulous annotations, and seamless integration, the RefSeq database emerges as a cornerstone in molecular research. Catering to a spectrum of scientific inquiries encompassing genomics, gene expression analyses, functional annotations, and myriad other investigative pursuits, it presents a compendium of reference sequences sourced from a broad array of organisms. This encompassing scope includes, though is not confined to, bacteria, archaea, plants, animals, fungi, and viruses.
RefSeq is a comprehensive repository housing reference sequences spanning genomes, transcripts, and proteins. Within the RefSeq database, stringent management methodologies are implemented to uphold the fidelity of gene sequences. Each sequence is accompanied by meticulous annotations, furnishing detailed insights into gene positions, exon/intron boundaries, alternative splicing isoforms, post-translational modifications, and functional domains. Drawing from a diverse array of resources, including empirical observations, computational predictions, and scholarly citations, RefSeq integrates data to offer dependable annotations for reference sequences.
Regular updates to the RefSeq database seamlessly incorporate emerging genomic, transcriptomic, and proteomic insights, guaranteeing scholars access to the most current reference sequences. This dynamic database fosters cross-referencing with other NCBI resources, such as GenBank and PubMed, empowering users to explore supplementary information and associated datasets in greater depth.
UCSC Genome Browser Database
The UCSC Genome Browser Database (https://genome.ucsc.edu/) stands as one of the most extensively utilized resources in the field of biology. Established and maintained by the University of California, Santa Cruz, it encompasses a wealth of genomic data, including gene annotation information (ENCODE), genome alignments, repetitive sequences, homologous sequences, reference sequences (mRNA, EST), phenotypes, expression profiles, regulatory information, conservation data, variations, and repetitive regions, among other information. UCSC encompasses genomic information for various common organisms, including humans, mice, fruit flies, zebrafish, nematodes, yeast, and others.
Moreover, it offers a suite of analysis tools to aid users in browsing gene information, accessing existing genome annotations, and downloading gene sequences. In the realm of bioinformatics analysis, the need for data files in formats such as fasta, GTF, or BED is ubiquitous, and UCSC serves as a primary source for accessing these files. Notably, Hg19 serves as a widely utilized reference sequence for the human genome within UCSC’s repertoire.